Regarding Extraction of Electric Field in Nerve Regions
-
I would like to extract the magnitude of the electric field only in the nerve region when a voltage is applied to the arm, based on the manual section 3.6.3 “Yoon-Sun Arm Stimulation,” using MATLAB. How should I approach this?
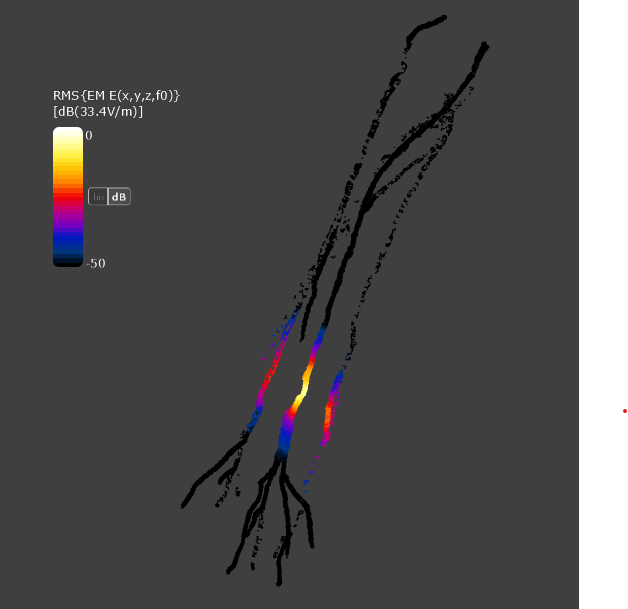
Also, when I try to visualize the electric field only in the nerve region using a mask filter (setting everything except Nerve_〇〇 to 0), the result appears discontinuous, as shown in the attached image. For the grid, I have set the maximum step and geometry resolution to 0.5 mm for all directions.
-
if it is discontinuous, then the voxel resolution was not sufficient to voxel the thinnest region of the nerve.
you can enforce continuous voxeling by dragging the nerve trajectories (splines/polylines) to the material settings (assign to "Nerve" material) and voxeler settings (assign a higher priority). The voxeler will assign all voxels intersected by the trajectories to "Nerve".
Warning: Note that for coarse voxels this will exaggerate the diameter of the nerve.
-
@bryn
Thank you for your reply.I followed the tutorial and set a high priority for the nerve region, but the visualization still appears discontinuous. However, when I applied a mask that sets all regions except the “nerve” and “neuron” to zero, the field became thicker but displayed smoothly, so I will proceed with this approach for now. (If there are any other possible improvements, I would appreciate your advice.)
Regarding the extraction of the electric field only in the nerve region as a .mat file — is this possible?
I tried dragging “nerve_〇〇” into the “overall field” to extract the electric field, but I received an error message saying “EM E contains a license protected geometry.”
According to what I found, this seems to mean that the electric field extraction is restricted due to license protection.
Could you please advise how this restriction can be avoided, or if there is an alternative way to export the field data only for the nerve region? -
great. yes, it makes sense that you need to specify the Nerve and the nerve trajectories in the masking filter.
Regarding the export to .mat file: I guess you could access the masked field as a numpy array in Sim4Life's Python and process/plot/export it from there, see e.g., this Python for MATLAB users cheatsheet
https://numpy.org/doc/stable/user/numpy-for-matlab-users.html
To access the masked field in Python you could use a script like this
import numpy as np import s4l_v1.analysis as analysis import s4l_v1.document as document import s4l_v1.model as model import s4l_v1.units as units import XPostProcessor as xp from s4l_v1 import Unit try: # Select your simulation by name here simulation = document.AllSimulations["LF - Copy"] simulation_extractor = simulation.Results() em_sensor_extractor = simulation_extractor["Overall Field"] em_sensor_extractor.FrequencySettings.ExtractedFrequency = u"All" inputs = [em_sensor_extractor.Outputs["EM E(x,y,z,f0)"]] field_masking_filter = analysis.core.FieldMaskingFilter(inputs=inputs) field_masking_filter.UpdateAttributes() # note I used a dummy simulation with a Nerve entity and a CenterLine spline, which I voxeled with the Nerve material properties nerve_entities = [model.AllEntities()["Nerve"].Id, model.AllEntities()["CenterLine"].Id] for uid in field_masking_filter.MaterialIds(): select = uid in nerve_entities field_masking_filter.SetMaterial(uid, False) field_masking_filter.Update() masked_field = field_masking_filter.Outputs[0].Data assert isinstance(masked_field, xp.FieldData) masked_field_array = masked_field.Field(0) assert isinstance(masked_field_array, np.ndarray) masked_field_grid = masked_field.Grid assert isinstance(masked_field_grid, xp.RectilinearGrid) print(masked_field.Quantity.Name) print(masked_field.Quantity.Unit) print(masked_field_array.shape) except Exception as exc: import traceback traceback.print_exc() raise(exc) -