Medical Image Segmentation
-
I faced a problem about segmentation by means of iSeg in sim4life light. I downloaded "iSeg_v1.1.zip" from "https://github.com/ITISFoundation/osparc-iseg/releases" and extracted it. Then, I chose FILE | Preferences settings of Sim4Life light and specified the correct path for iSeg.exe in my computer. I imported dicom medical image data successfully. In the ribbon, I clicked "Image Tools|Segmentation Group". This generated en empty LabelField entity. Then, I
selected the LabelField item in the Explorer window and, in the ribbon, "iSeg" icon was not available to start this software and segment images. The attached figure demonstrates my problem better.
Thank you for your reply.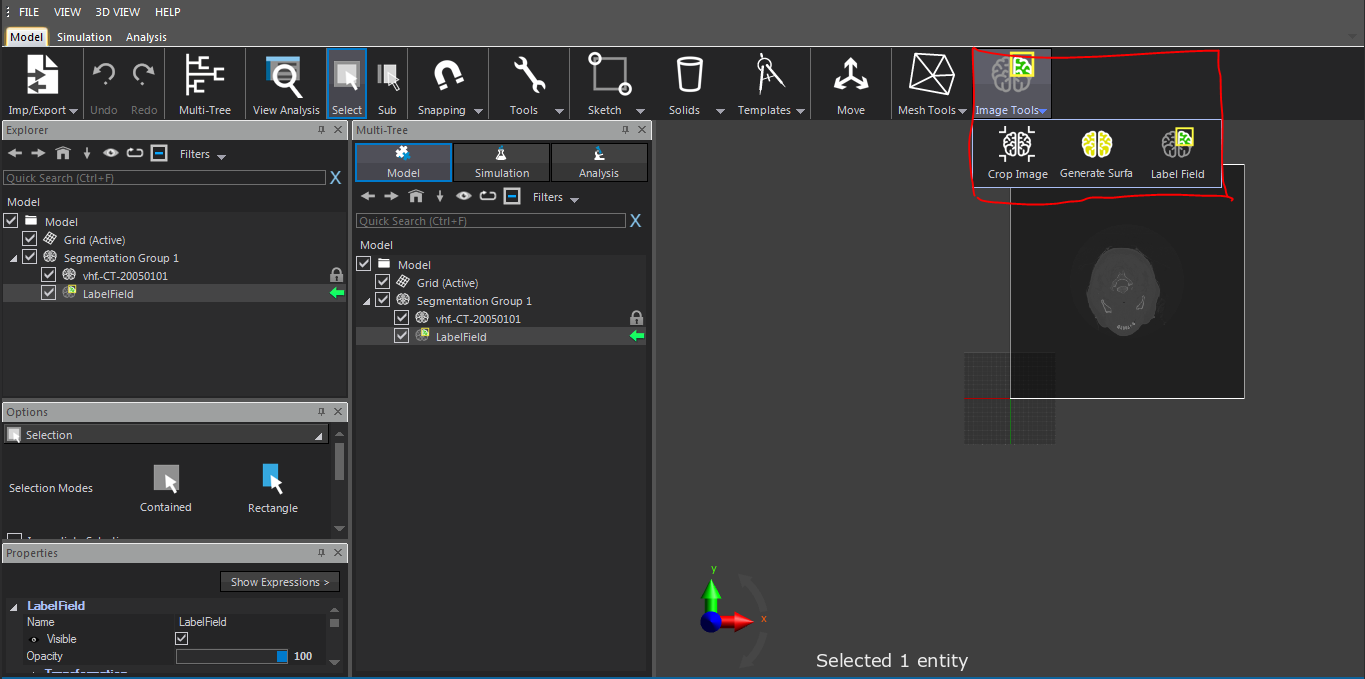
-
The link between Sim4Life and iSeg is a licensed tool and is not supported by Sim4Life light. The commercial/academic version of iSeg has to be used in this case.
For the open source version, you have the option to export the segmented images to a format that can be imported into Sim4Life (e.g. STL or VTK)
-
To comment on this case. The surface extraction in iSEG is inferior to the surface extraction in Sim4Life. So you probably want to export the labelfield as voxels (e.g. in Nifty, Nrrd or Metaheader format - the VTK format does not preserve the transformations correctly). You can also extract the tissue list as txt file in iSEG in order to set the tissue names e.g. via a python script in Sim4Life.