IMAnalytics Data Preprocessing
-
Hi,
I am looking to use IMAnalytics to extract deposited power on lead pathways I drew within the human body models (Fats model). Is this tool only usable with the MRIxViP field libraries? or can I use field data I have simulated too?
I'm wondering how I can generate the 'JSON' preprocessed index file for my simulation setup. A section of the manual went over doing this using MRIxViP field library. What would be the difference in the process if I am using my own field data. Here is that section in the manual:
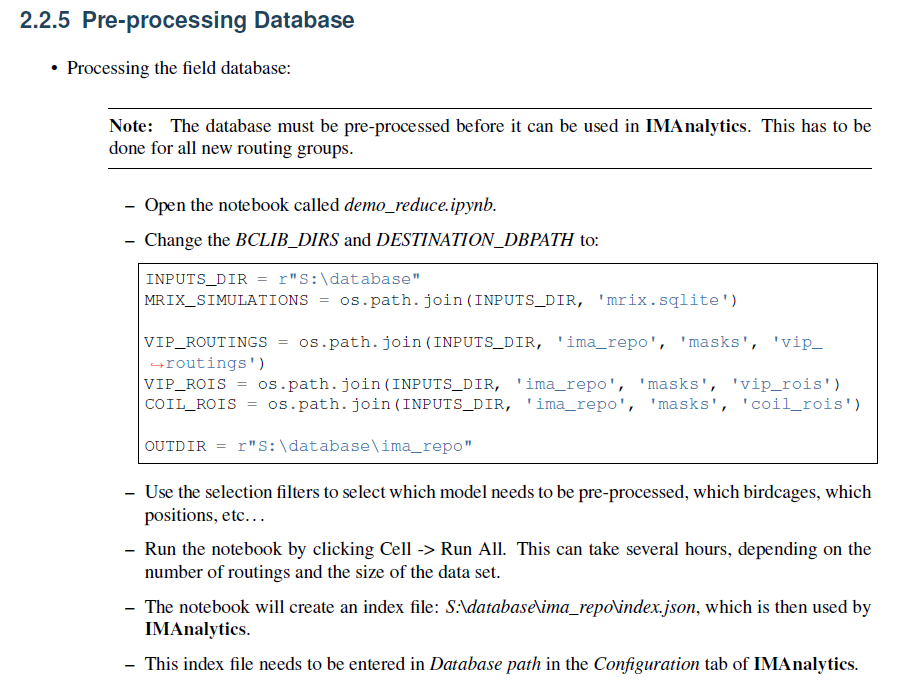
I appreaciate the help!
Thanks :beaming_face_with_smiling_eyes:
-
Hi,
I am looking to use IMAnalytics to extract deposited power on lead pathways I drew within the human body models (Fats model). Is this tool only usable with the MRIxViP field libraries? or can I use field data I have simulated too?
I'm wondering how I can generate the 'JSON' preprocessed index file for my simulation setup. A section of the manual went over doing this using MRIxViP field library. What would be the difference in the process if I am using my own field data. Here is that section in the manual:

I appreaciate the help!
Thanks :beaming_face_with_smiling_eyes:
Hi @mkn ,
The input of IMAnalytics is a database of simulation results. The MRIxViP field library is one such database but any similar field database will work too provided that:
- the simulation results are output files from Sim4Life simulations
- the output files are organized in a folder structure that matches that of the MRIxViP field library: e.g. "64MHz\HP_B70_L60\Duke\HP_B70_L60_64_Duke_V3_350_Results.smash\a17e7f8e-e520-4f08-af16-1aafc1e1b830_Output.h5", where "64", "HP_B70_L60", "Duke", "Duke_V3" and "350" indicate the frequency, coil name, model name, model name with version, and landmark position, respectively.
- the output files contain a field sensor named "Field Sensor Duke" (if the model is called Duke)
- the output files contain no other field sensors
- the simulation name with which each output file is created is named following the pattern "HB_{channel_name}{model_name_with_version}{position}", e.g. "HB_Q_Duke_V3_350" for a simulation at position 350 of Duke with the Q channel excited. Most important is that the channel name is exactly after the first underscore ("_") character.
There is a notebook in the examples of IMAnalytics that indexes this database. It is then possible to inspect this database programmatically to ensure that it is 100% complete and accurate - admittedly not an easy task, depending on how many simulations need to be integrated, but perfectly doable with the Python tools at hand.
Note that, before you are able to create the 'JSON' preprocessed index file, you will have to first index the field library itself (this creates the file named
mrix.sqlite). You can find an example of this in the notebook calledindexing_mrx.ipynbFuture versions of IMAnalytics will relax some of these constraints on the format and structure of the field library but the next release date has not yet been fixed.
I hope this helps.
-
Hello @Sylvain,
Thanks for your response! This points me in the right direction.
My results were actually simulated in SEMCAD 14.x, but I'm guessing it can easily be imported into Sim4life for postprocessing. The main thing I am concerned about is the difference in file naming and so on that may not be the same as the MRIxViP field library? Can this be modified without invalidating the results altogether?
Another question I have is regarding I and Q channel sweeping, for the time being I'm only interested in the case of circular polarization (not sweeping I and Q). I'm guessing this shouldn't be an issue?
As for the number of simulations at hand, for now I am only considering a single simulation with a single birdcage and a few positions, with a number of pathways included for evaluation.
Thank you again for your help!
-
Hi @mkn,
I actually don't know for sure how easy it would be to import your SEMCAD 14.x results into Sim4Life while changing the simulation names in such a way that they are correctly written into the results files (those ending with Output.h5). Timewise, it's probably simpler to just re-run the simulations with compatible names...
If you are only interested in a handful of simulations, you might consider using the IMSAFE tool of Sim4Life instead of IMAnalytics. IMSAFE is designed to extract the tangential E-field values along different pathways and lets you export the data as plain-text files. It does not perform the convolution with the transfer function (to evaluate deposited power and/or induced voltage), but maybe that is sufficient for your needs? IMAnalytics is geared towards large simulation databases, so it might feel over-engineered if you only have few simulations.
As for channel sweeping, IMAnalytics lets the user choose the list of excitation vectors that need to be considered for the evaluations. The list could very well consist of a single vector, corresponding to the circular polarization.
-
Hi @Sylvain,
Thanks for your replies and help! I was able to import the SEMCAD file for one of the HBM into Sim4Life, took sometime but it's doable (less time than having to re-simulate everything, and the simulation name can be changed to match the format you suggested earlier "HB_Q_Duke_V3_350". Here are the output file names generated,
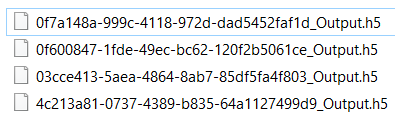
It seems correct to me, would this work fine to move further with indexing?
I have started using IMSafe as well. My plan is to test IMAnalytics with a single model to ensure it will work with my data then incorporate more models later on.
Now I have two questions:
-
You mentioned there is a notebook example to index the database to match that of the MRIxVIP, my understanding is that this will take the Sim4Life results in the .smash file and add the "output" files in the correct folders to match that of the MRIxVIP library. Is this correct?
-
350 in "HB_Q_Duke_V3_350" refers to the position, is this the position relative to a given coordinate system? What I mean is the coil/body is moved 350mm from origin?
Thank you!
-
-
hi @mkn,
I am not sure the indexing will work correctly following your import+renaming workflow. The reason (and that will answer your first question too) is that the indexing is done directly on the 'Output.h5' files and completely ignores the .smash ones. So renaming the simulations in the Sim4Life project may not have the desired effect, unless it also modifies the Output.h5 results.
The indexing also does not move, modify or otherwise reorganize your collection of Output.h5 files. This has to be done manually, following the folder hierarchy that I described earlier. The indexing itself simply parses the files it finds and builds an index (ending with .sqlite). It is basically at this stage that your output.h5 files acquire some semantic "meaning" (i.e. that they are associated with a given birdcage coil, anatomical model, etc...).The "position" is indeed the relative position of the body with respect to the coil. In MRIxViP, the landmark position is defined as the distance between the isocenter of a given bone in the skull of the body and the isocenter of the coil, along the direction of the z-axis of the coil. Note that IMAnalytics will blindly trust that the Output.h5 files it finds in a given folder correspond to the configuration determined by the indexing process (i.e. the labelling of the data is done by placing the files in the correct folders).