Padding field with constant value
Analysis & Postprocessing
1
Posts
1
Posters
494
Views
1
Watching
-
Recently a user asked how they could extract a closed surface from a field, e.g. at the iso-value contour of a field distribution.
I answered that you can extract the iso-contour surface using the IsoSurfaceViewer.The problem was then that the iso-surface can be open, if the iso-value passes through the boundary. So I suggested to pad the field with a constant value (e.g. very small value) first, forcing the contour surface to be closed at the border.
But how can you pad a field using the Sim4Life api? Since this may be useful/interesting for other users I would like to share a snippet showing how this can be achieved:
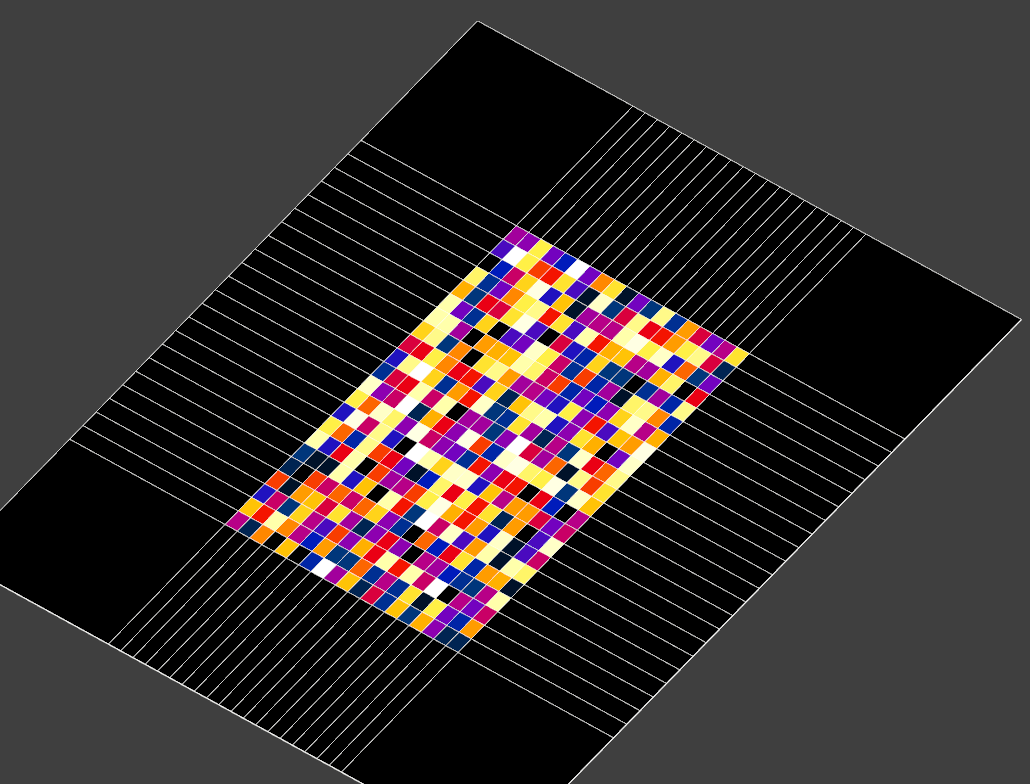
import s4l_v1 as s4l import s4l_v1.document as document from s4l_v1 import Unit import numpy as np def create_rectilinear_field(nx: int, ny: int, nz: int, num_components: int = 1, cell_data: bool = True, value: float = 1.0): def make_quantity(name: str, unit: str): quantity = s4l.analysis.core.Quantity() quantity.Name = name quantity.Unit = Unit(unit) return quantity grid = s4l.analysis.core.RectilinearGrid() grid.XAxis = np.array(np.linspace(-0.15, 0.15, nx)) grid.YAxis = np.array(np.linspace(-0.25, 0.25, ny)) grid.ZAxis = np.array(np.linspace(-0.3, 0.3, nz)) grid.Unit = Unit("m") num_tuples = (nx - 1) * (ny - 1) * (nz - 1) if cell_data else nx * ny * nz location = s4l.analysis.core.eValueLocation.kCellCenter if cell_data else s4l.analysis.core.eValueLocation.kNode field_data = s4l.analysis.core.FloatFieldData() field_data.Grid = grid field_data.ValueLocation = location field_data.Allocate(1, num_tuples, num_components) field_data.SetField(0, np.random.rand(num_tuples, num_components)) field_data.Quantity = make_quantity("EM E(x,y,z,f0)", "V/m") field_data.SnapshotQuantity = make_quantity("Frequency", "Hz") field_data.Snapshots = [1000] return field_data def pad_rectilinear_field(field_data: s4l.analysis.core.FieldData, pad_width: int = 1, value: float = 0.0, dx: float = 1e-3): grid = field_data.Grid assert isinstance(grid, s4l.analysis.core.RectilinearGrid) grid_padded = grid.Clone() grid_padded.XAxis = np.pad(grid.XAxis, pad_width=pad_width, mode="linear_ramp", end_values=(grid.XAxis[0]-dx, grid.XAxis[-1]+dx)) grid_padded.YAxis = np.pad(grid.YAxis, pad_width=pad_width, mode="linear_ramp", end_values=(grid.YAxis[0]-dx, grid.YAxis[-1]+dx)) grid_padded.ZAxis = np.pad(grid.ZAxis, pad_width=pad_width, mode="linear_ramp", end_values=(grid.ZAxis[0]-dx, grid.ZAxis[-1]+dx)) data = field_data.Field(0) num_components = data.shape[1] nx, ny, nz = grid.XAxis.size, grid.YAxis.size, grid.ZAxis.size if field_data.ValueLocation == s4l.analysis.core.eValueLocation.kCellCenter: nx, ny, nz = nx - 1, ny - 1, nz - 1 data = np.reshape(data, (nz, ny, nx, -1)) data_padded = np.pad(data, pad_width=[(pad_width,), (pad_width,), (pad_width,), (0,)], mode="constant", constant_values=value) nx_padded, ny_padded, nz_padded = nz+2*pad_width, ny+2*pad_width, nx+2*pad_width data_padded = data_padded.reshape((-1, num_components)) field_padded = field_data.Clone() field_padded.Grid = grid_padded field_padded.Allocate(1, nx_padded * ny_padded * nz_padded, num_components) field_padded.SetField(0, data_padded) return field_padded if __name__ == "__main__": field = create_rectilinear_field(20, 23, 29, 1, True) field_padded = pad_rectilinear_field(field, 1, 0.0, dx=0.2) source = s4l.analysis.core.TrivialProducer() source.AddDataObject(field) source.AddDataObject(field_padded) document.AllAlgorithms.Add(source)This is what it would like in a slice field viewer:
before padding: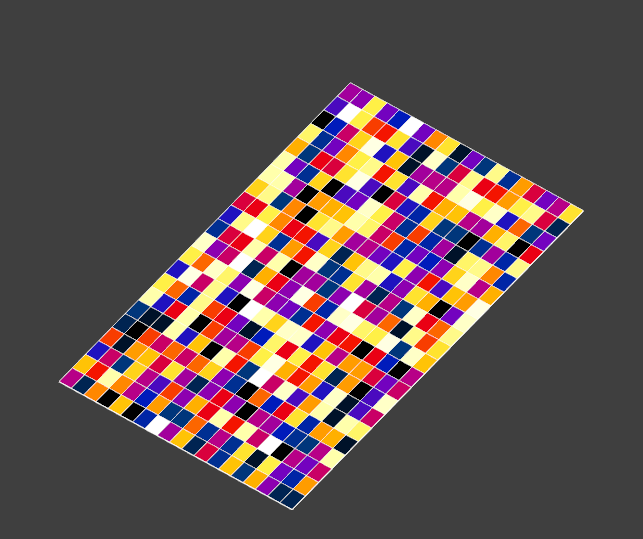
after padding: