Automatic Head Model including 1010-System / Electrode Placement
-
The T1w image is placed in world (scanner) coordinates. This is useful, e.g. to align different acquisitions (e.g. T1, T2 with different resolutions or field of view).
You could remove the rotation and translation, e.g., by setting an identity transform. However, in my experience, it is often useful to preserve the position in world coordinates.
img = XCoreModeling.Import("some_t1w_mri.nii.gz") img.Transform = XCoreModeling.Transform() -
The T1w image is placed in world (scanner) coordinates. This is useful, e.g. to align different acquisitions (e.g. T1, T2 with different resolutions or field of view).
You could remove the rotation and translation, e.g., by setting an identity transform. However, in my experience, it is often useful to preserve the position in world coordinates.
img = XCoreModeling.Import("some_t1w_mri.nii.gz") img.Transform = XCoreModeling.Transform() -
I don't understand what you are doing. The error looks like Sim4Life cannot parse the .pts file produced by the landmark predictor.
- did you edit the .pts file manually?
- where/how did you set the rotation/translation to "0"?
- did you try to set the transform [to zero] (before running the prediction) as suggested in my last post?
-
-
I can reproduce your issue. If I set the transform to "0", i.e. Identity, the predictor fails. The head40 segmentation is also less accurate! We need to investigate.
A workaround would be:
- load image
- predict landmarks & segmentation
- compute inverse image transform
- apply this inverse to landmarks/segmentation/surfaces/etc
# assumes verts and labelfield are already predicted (without setting transform to "0") inv_tx = img.Transform.Inverse() # transform segmentation labelfield.ApplyTransform(inv_tx ) # transform landmarks for v in verts: v.ApplyTransform(inv_tx ) -
The issue is that our neural network was trained with the data in the RAS orientation (with some deviation +- 15 degrees, and flipping in all axes). If you manually edit the transform, you break assumptions used to pre-orient the data into RAS ordering.
Since RAS is a widely used convention in neuroscience, and medical images are always acquired with a direction (rotation) matrix and offset (translation), I think it is best you don't modify the transform.
For instance, if you try to assign DTI-based conductivity maps - you will need to rotate the grid AND the tensors accordingly. It can be done, but it will be more effort...
If this is to investigate if the fields are (nearly) symmetric, I suggest you
- find an approximate symmetry plane (wrt to the brain or skull or ...)
- align the plane of a slice viewer perpendicular to the symmetry plane
-
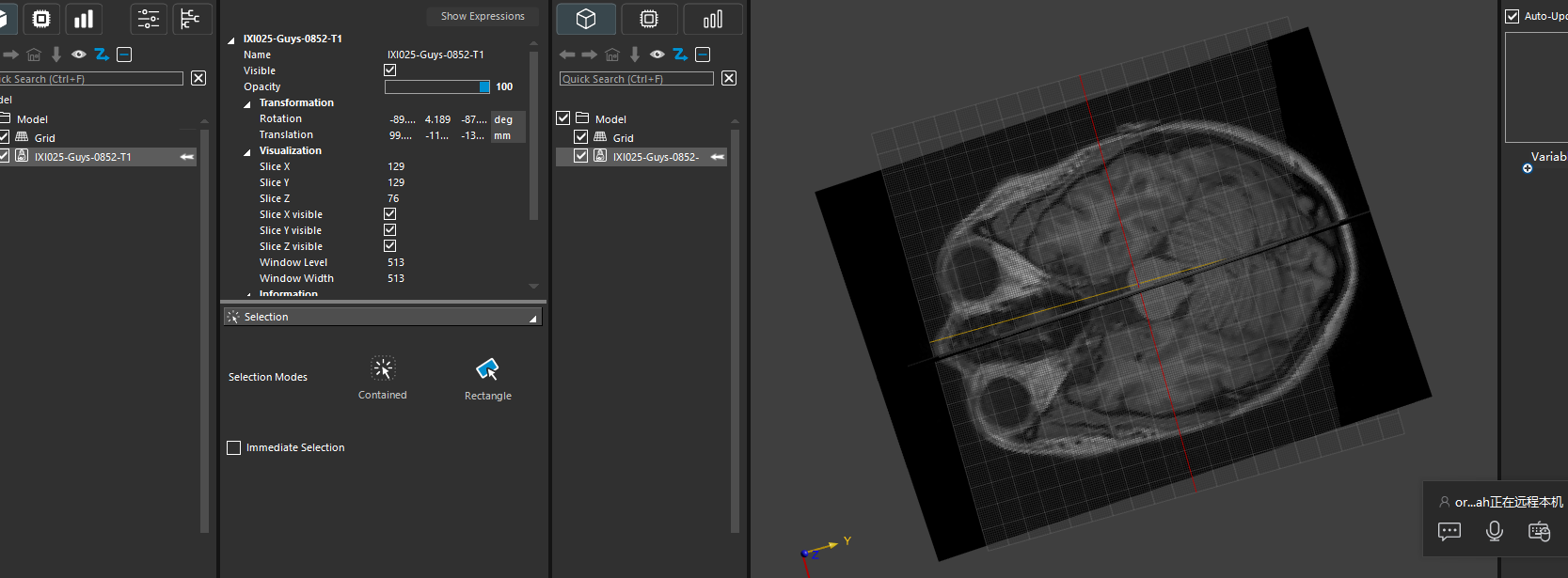
The latest release 8.2 includes a new function to predict the landmarks needed to place the 10-10-system on the skin:
Predict1010SystemLandmarks
The landmarks are the nasion, inion, and left/right pre-auricular points. Sim4Life now can predict these directly from a T1w MRI.The following script demonstrates the whole process:
from ImageML import Predict1010SystemLandmarks from s4l_v1.model import Vec3, Import, Create1010System, PlaceElectrodes, CreateSolidCylinder from s4l_v1.model.image import HeadModelGeneration, ExtractSurface img = Import(r"D:\datasets\IXI-T1\IXI021-Guys-0703-T1.nii.gz")[0] # segment head, skip adding dura, labelfield = HeadModelGeneration([img], output_spacing=0.6, add_dura=False) # extract surfaces from segmentation surfaces = ExtractSurface(labelfield) surfaces_dict = {e.Name: e for e in surfaces} skin = surfaces_dict["Skin"] # predict landmarks, the function returns a list of Vertex entities verts = Predict1010SystemLandmarks(img) pts = {e.Name: e.Position for e in verts} eeg1010_group = Create1010System(skin, Nz=pts["Nz"], Iz=pts["Iz"], RPA=pts["RPA"], LPA=pts["LPA"]) eeg1010_dict = {e.Name: e for e in eeg1010_group.Entities} # create template electrode and place it at C3 position electrode_template = CreateSolidCylinder(Vec3(0), Vec3(0,0,5), radius=10) electrodes = PlaceElectrodes([electrode_template], [eeg1010_dict["C3"]])For the image used in this example, the result looks like this:

-
The default is 40 tissues. To be explicit you can specify this via
import ImageML labelfield = ImageML.HeadModelGeneration([img], output_spacing=0.6, add_dura=False, version=ImageML.eHeadModel.head40)For 30 (or 16) tissues you would specify the version
head30(orhead16)import ImageML labelfield = ImageML.HeadModelGeneration([img], output_spacing=0.6, add_dura=False, version=ImageML.eHeadModel.head30)But please note: the versions are an evolution. The
head16segmentation is not the same, with fewer tissues. It is also less accurate, as it was the first version we published (and trained on less training data). -
The default is 40 tissues. To be explicit you can specify this via
import ImageML labelfield = ImageML.HeadModelGeneration([img], output_spacing=0.6, add_dura=False, version=ImageML.eHeadModel.head40)For 30 (or 16) tissues you would specify the version
head30(orhead16)import ImageML labelfield = ImageML.HeadModelGeneration([img], output_spacing=0.6, add_dura=False, version=ImageML.eHeadModel.head30)But please note: the versions are an evolution. The
head16segmentation is not the same, with fewer tissues. It is also less accurate, as it was the first version we published (and trained on less training data).@bryn Thank you very much for your response! I have a question: What is the difference between constructing a head model using T1-weighted (T1W) and T2-weighted (T2W) images and constructing a head model using only T1W images? Why can only 16 types of tissues be segmented when using T1W and T2W images?
-
@bryn Thank you very much for your response! I have a question: What is the difference between constructing a head model using T1-weighted (T1W) and T2-weighted (T2W) images and constructing a head model using only T1W images? Why can only 16 types of tissues be segmented when using T1W and T2W images?
@lucky_lin In our first version of the head segmenation (head16) we trained with a smaller dataset, where T1w and T2w was available. We trained two networks, one with just T1w as input, one that gets T1w + T2w as input.
In our later work we extended the training data, but only have T1w images. Therefore, the head30 and head40 only needs a T1w image.
-
@lucky_lin In our first version of the head segmenation (head16) we trained with a smaller dataset, where T1w and T2w was available. We trained two networks, one with just T1w as input, one that gets T1w + T2w as input.
In our later work we extended the training data, but only have T1w images. Therefore, the head30 and head40 only needs a T1w image.
-
The latest release 8.2 includes a new function to predict the landmarks needed to place the 10-10-system on the skin:
Predict1010SystemLandmarks
The landmarks are the nasion, inion, and left/right pre-auricular points. Sim4Life now can predict these directly from a T1w MRI.The following script demonstrates the whole process:
from ImageML import Predict1010SystemLandmarks from s4l_v1.model import Vec3, Import, Create1010System, PlaceElectrodes, CreateSolidCylinder from s4l_v1.model.image import HeadModelGeneration, ExtractSurface img = Import(r"D:\datasets\IXI-T1\IXI021-Guys-0703-T1.nii.gz")[0] # segment head, skip adding dura, labelfield = HeadModelGeneration([img], output_spacing=0.6, add_dura=False) # extract surfaces from segmentation surfaces = ExtractSurface(labelfield) surfaces_dict = {e.Name: e for e in surfaces} skin = surfaces_dict["Skin"] # predict landmarks, the function returns a list of Vertex entities verts = Predict1010SystemLandmarks(img) pts = {e.Name: e.Position for e in verts} eeg1010_group = Create1010System(skin, Nz=pts["Nz"], Iz=pts["Iz"], RPA=pts["RPA"], LPA=pts["LPA"]) eeg1010_dict = {e.Name: e for e in eeg1010_group.Entities} # create template electrode and place it at C3 position electrode_template = CreateSolidCylinder(Vec3(0), Vec3(0,0,5), radius=10) electrodes = PlaceElectrodes([electrode_template], [eeg1010_dict["C3"]])For the image used in this example, the result looks like this:

-
@lucky_lin In our first version of the head segmenation (head16) we trained with a smaller dataset, where T1w and T2w was available. We trained two networks, one with just T1w as input, one that gets T1w + T2w as input.
In our later work we extended the training data, but only have T1w images. Therefore, the head30 and head40 only needs a T1w image.