voxel of individual anatomic model creates holes
-
I created my own anatomic model based on a patient CT data set. I have different Roi´s made by a doctor and fat, muscle and bone is created with threshholding software. At the end of process i have a anatomic voxel model without any empty spaces ( double checked it with "Amira"-software and iSeg) I import my model to Sim4Life and do the voxeling process. After that I get irregular holes ( properties like the background) in my model. Small and big holes in different Roi´s, not only in every slice. I tested a smaller and a bigger Gridsize and other models, same problems.
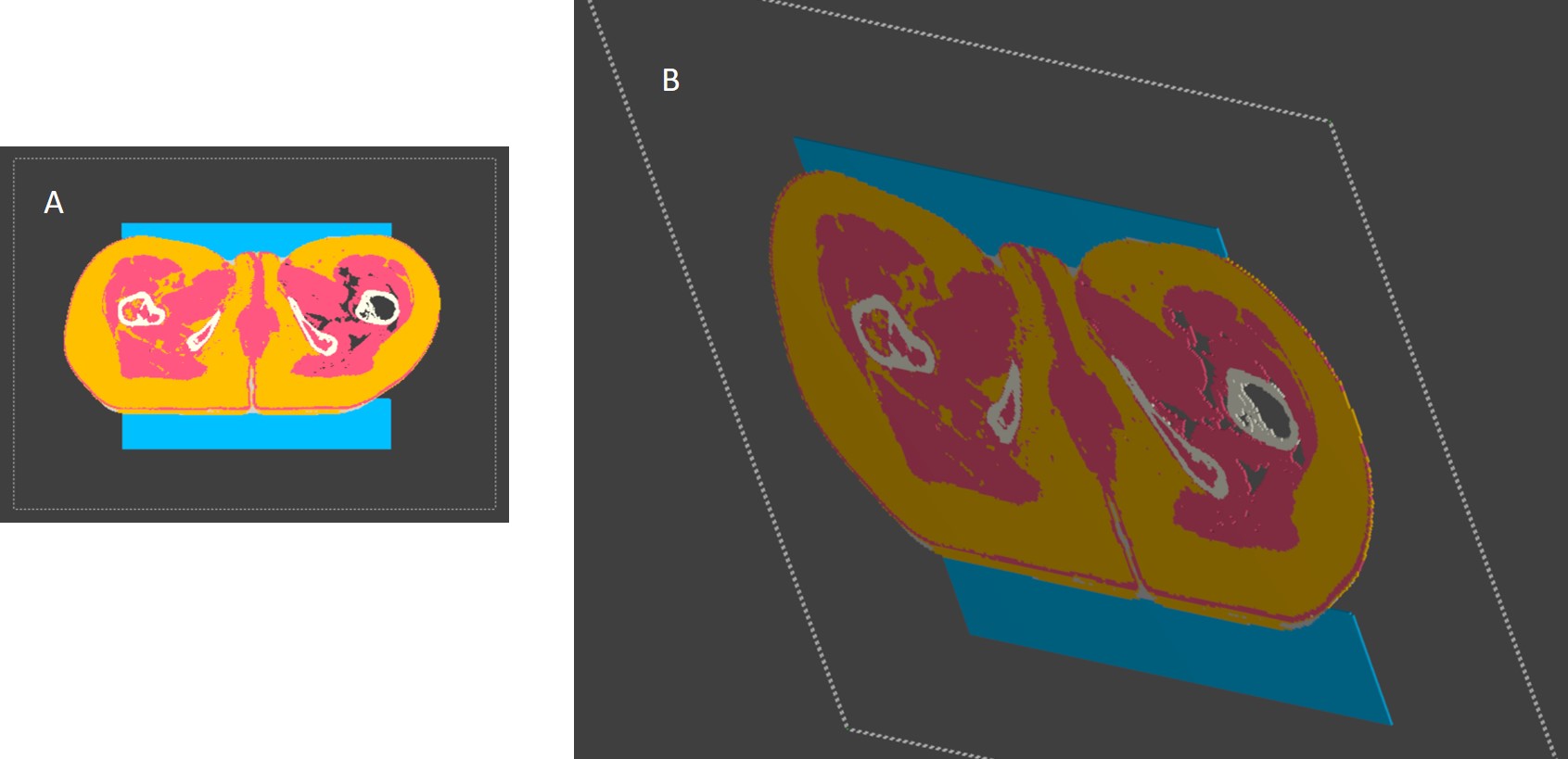
Is there a for my problem? Or is it possible to load only voxels without the process "create triangles to meshes"?Pic A There is no Fat (yellow in some regions). you can see that there are no Voxels (pic B)
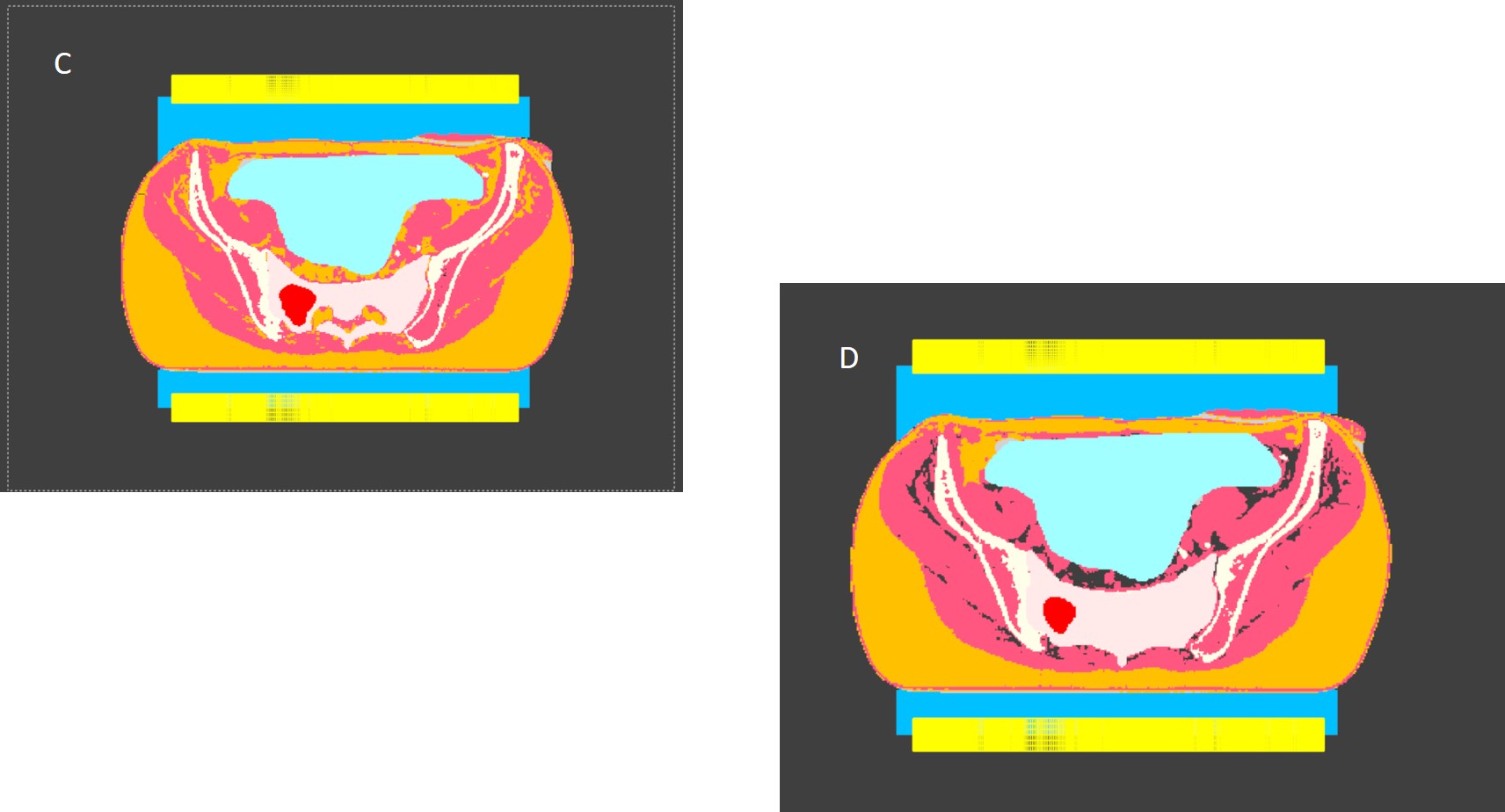
Pic C Everything is filled with Fat (yellow), Muscle (red) Bone (white) and doctors´s Structures, but in the next slice (Pic D) there are holes (grey)
-
How do you import the segmentation into Sim4Life: By loading the iSEG project file (*.prj) or via Import Voxels (with the option "Create Solids" on or off)?
If you are importing voxels, I would recommend to select "Create Solids" = OFF. Then, since you cannot directly assign the imported label field to a simulation, generate a surface-based model (Image Tools -> Generate Surfaces).
Also, try to avoid create surfaces of different tissues separately, or post-processing them separately, if possible. The Generate Surfaces tool creates a solid model with no holes, gaps or overlaps.
I cannot guess where the issue in C/D is coming from, except that the voxeler is having problems with the solids (if you selected "Create Solids"=ON in the voxel import dialog).
-
Has anyone solved/worked on this issue since?
I have also found the work-around by not creating solids, but this also means that I lose the information of region names and priorization which would have been included in the descriptor-file. All my regions are now named "tissue XX" and are imported in an order differing from the label number in the raw file.I'd appreciate any help / news on this!
-
-
If you provide a tissue descriptor file, tge tissue names should not get lost. This is something I have used frequently, so please try again and check if there was a problem importing the descrptors in the console
-
If i get the context right, you are importing raw voxels. A voxel has only one value, so there are no label overlaps and no priorities. As far as i remember the labels are not reordered. Maybe check how you generate the voxels
-
-
I now realize I was missing information by mixing two (unrelated?) problems I have:
- If I try to use the descriptor file (not manual mode), with or without creating solids, nothing happens but I also don't get any error message in the log.
- When I then move to manual mode, I get one of the following:
a) I check "Create solids" and use the descriptor file under "use color names" -> tissue name and order are as intended but I get the same holes in the voxel model Tom described in the post opening this thread.
b) I don't create solids and end up with wrong tissue names and an import order that differs from the label order which makes it very cumbersome to assign the regions afterwards. (Feasible, of course, but time consuming.)
The descriptor file is formatted as indicated in the documentation, resolution and grid size are the same I use in manual mode (= are correct) and when I use the file for "coloring", names and regions match.
Since I don't get any error messages, I don't understand what the problem is and cannot use the workaround of using the deescriptor file without creating solids.
-
I am on vacation atm, but could debug what is going on with your data when I am back (if you are able to send it).
As a workaround you could save the voxels in a medical image format, like nifti (*nii, *nii.gz), nrrd or metaheader (mhd, mha) and import the file as labelfield, see also this discussion: https://forum.zmt.swiss/topic/270/importing-file-as-a-label-field
Assuming you have the segmentation loaded (without reordering), you can also assign the tissue names AFTER importing, via the Python API.
TheLabelFieldentity has the methodsLoadTissueListandSaveTissueList. To understand the format you can first save the current tissue list. The format is the same as used by iSEG, and very simple. The first line is the version number, second line contains number of tissues, then each line (skipping label=0) is listed (color as RGBA and name - the alpha value is ignored). Here is an example:V7 N15 C0.00 0.80 1.00 0.50 Air_internal C1.00 0.40 0.00 0.50 Artery C0.00 0.40 1.00 0.50 Bone_cancellous C1.00 0.80 0.00 0.50 Bone_cortical C0.00 0.00 1.00 0.50 Cerebrospinal_fluid C0.80 1.00 0.00 0.50 Cerebrum_grey_matter C0.40 0.00 1.00 0.50 Cerebrum_white_matter C0.40 1.00 0.00 0.50 Eyes C0.80 0.00 1.00 0.50 Mucosa C0.00 1.00 0.00 0.50 Other_tissues C1.00 0.00 0.80 0.50 Rectus_muscles C0.00 1.00 0.40 0.50 Skin C1.00 0.00 0.40 0.50 Spinal_cord C0.00 1.00 0.80 0.50 Vein C1.00 0.00 0.00 0.50 Visual_nerve -
D dbsim4 referenced this topic on